Flow cytometry technique for identification and functional analysis of NK cells
- Post by: wsmib-admin
- septembre 17, 2024
- Comments off
Flow cytometry technique for identification and functional analysis of NK cells
Simfele Hombamane Christelle1*, Nadjir Koboyo Liza2, Kolou Malewe3, Katawa Gnatoulma1
1Unité de Recherche en Immunologie et Immunomodulation (UR2IM) /Laboratoire de Microbiologie et
de Contrôle de Qualité des Denrées Alimentaires (LAMICODA)/Ecole Supérieure des Techniques
Biologiques et Alimentaires (ESTBA), Université de Lomé, Lomé, Togo
2Centre National de Transfusion Sanguine (CNTS), Lomé, Togo
3Faculté des Sciences de la Santé, Université de Lomé, Lomé, Togo
Abstract
Introduction
Natural killer (NK) cells are lymphocytes subset that contribute to innate immunity through their cytotoxic function. This cytotoxic function is generally provided by degranulation of CD56dimCD16pos NK subpopulation during various infections. However, the identification of NK cells can be complex due to their different functions. Here we propose a protocol for the identification of CD56dimCD16pos NK cells by flow cytometry.
Methods
Peripheral blood mononuclear cells (PBMCs) from healthy individuals were isolated by density gradient centrifugation using Ficoll-Paque; then NK cell subsets and degranulation markers were characterised by flow cytometry using Annexin-V, CD3, CD56, CD16 and CD107a staining.
Results
After showing the gating strategies for the identification of unstained and fluorescence minus one (FMO) control, the different NK cell subpopulations will be displayed: CD56negCD16pos, CD56dimCD16pos, CD56brightCD16dim, CD56dimCD16neg and CD56brightCD16neg. Finally, the gating strategy for the functional analysis of the degranulation of the CD56dimCD16pos subpopulation will be presented: CD56dimCD16posCD107apos.
Conclusion
This protocol will facilitate the functional analysis and identification of degranulated NK cells during various infections.
Keywords
Natural killer (NK) cell subsets, Peripheral blood mononuclear cells, Degranulation, Flow cytometry
Introduction
Natural killer (NK) cells are granular subset of lymphocytes that are part of the innate immune system. They play a crucial role in the initial defence against viral, bacterial or parasitic infections (1). In humans, NK cells are CD3 negative cells and constitute about 5-15% of peripheral blood mononuclear cells (PBMCs), and they can typically be identified by the expression of the surface markers CD56 and
CD16 (2, 3). There are two main subsets of NK cells with distinct maturation and functional properties (4).
CD56BrightCD16neg subsets are efficient producers of pro-inflammatory cytokines and represent 10% of the peripheral NK cells (5). In contrast, CD56dim CD16pos NK cells represent about 90% of peripheral NK cells in humans and are known for their high cytotoxic effector properties (6). Some reports have described a subset of CD56neg CD16pos NK cells that are expanded in the context of chronic viral infections such as Human Immunodeficiency Virus (HIV) patients, (7) or during Hepatitis C Virus (HCV)
infections (8). Regarding the cytolytic function of NK cells, the most abundant protein in the cytotoxic granules is CD107a, and its expression on the surface is considered as a marker of degranulation (9). Due to the important role that NK cells play in immunity, and the emerging exploration of the NK cell platform for immunotherapy in different diseases, analysis of NK cell responses is becoming a useful tool for assessing a variety of clinical conditions, such as cancer, post-stem cell transplantation to assess risk of
infection and diagnosis of immunodeficiency syndromes. However, the identification of NK cells can be complex due to their different functions. Here we propose a simple and easy method for identifying the different subpopulations of NK cells and analysing their cytotoxic function by flow cytometry.
Methods
Materials
All materials used in this protocol are listed in the Table 1.
Buffer and media preparation
Fetal calf serum preparation
Inactivate Fetal Calf Serum (FCS) complement with humid heat for 30 minutes at 56°C before use.
RPMI 1640 Complete Medium preparation
To 500mL RPMI 1640, add 10% inactivated FCS supplemented with Gentamycin at a concentration of
50µg/mL, Penicillin/Streptomycin at a concentration of 100µg/mL and L-glutamine at a concentration of 2 mM/mL.
Cell Cryopreservation Medium (Freezing medium)
Add 20% DMSO to the inactivated FCS to obtain the freezing medium. Then chill the medium for 5 to 10 minutes to limit the exothermic reaction that occurs when DMSO is added to FCS, before adding it to the cells.
FACS buffer preparation
To sterile PBS add 2% of inactivated FCS to obtain the FACS Buffer.
Peripheral Blood Mononuclear Cells (PBMCs) Isolation
PBMCs from 3 healthy donors were isolated using the Ficoll Paque density gradient centrifugation method.
- Collect 20mL of blood by vein-puncture in EDTA tubes from each participant as followed:
- Add 10 mL of Ficoll-Paque to three conical 50mL tube.
- Dilute the whole blood at 1:2 with sterile PBS in a 50mL conical tube and mix gently. In this protocol
we have used 10mL of PBS (1x) and 20mL of whole blood. - Carefully transfer the diluted blood sample on the top of Ficoll-Paque into each 50 mL conical tube
avoid mixing the Ficoll-Paque and the diluted blood. - Centrifuge tubes at 2000×g for 20 minutes at room temperature.
- Collect the white interphase layer of PBMCs with a sterile pipette in a 50mL conical and wash twice by centrifugation at 1300 rpm for 8 minutes with 10mL RPMI 1640 medium previously prepared.
- Resuspend the cells pellet in 1mL of complete medium.
- Count viable cells by Trypan Blue exclusion technique.
- If needed, add the cells suspension with RPMI 1640 medium to obtain 1×105 cells/mL.
NB: The entire isolation procedure must be carried out under a hood and the cells must be kept on ice.
PBMCs cryopreservation
The rest of the fresh cells can be preserved for further use.
- Prepare cell cryopreservation medium.
- We usually freeze 1×105 cells per cryovial.
- Aliquot into 2mL cryotubes a total volume of 500µL of cells each containing 500µL of freezing
medium to obtain a final volume of 1mL. - Place all the vials in a freezing container and store it at −80°C. Then, transfer the vials to an appropriate container and store them in liquid nitrogen.
Note: The cryopreservation process needs to be fast. As it affects cell viability, try to shorten the time that cells are exposed to DMSO before putting them in the freezer.
Immunophenotyping of NK cells
The surface staining and fixation procedures are described in detail in this step.
Use fresh PBMCs for phenotypic and functional assays. To assess NK cells degranulation, a surface staining can be performed.
- Prepare the tubes for NK cells immunophenotyping
as followed:
a. Unstained tube: negative control.
b. Fluorescence minus One (FMO) tubes: FMO control contains all the fluorochromes in a panel, except for the one that is being measured.
c. Tube test: it contains all the fluorochromes in a panel. - Pipette 100μL PBMCs suspension containing 1×105 cells in each tube.
- Wash the cells, by centrifugation at 1500 rpm for 5 minutes with 200μL of FACS buffer and discard the supernatant.
- Stain the cell pellets in the dark with appropriate markers in appropriate tube, following the
manufacturer’s instructions. In this protocol we used anti-hCD3-APCA750; anti-hCD16-FITC and
anti-hCD56-PE as specific markers of NK cells ; anti-h Annexin-V PB450A for viability staining;
anti-hCD107a APC for degranulation assays. Vortex the stained cells and incubate under dark for
30 minutes at 4°C. - Repeat the step 3 and resuspend the cells in 100 µL of FACS buffer for sample acquisition on the flow cytometer.
Note: Alternatively, you can also prepare one control per marker that you want to determine (Single stained) to assess the effectiveness of markers.
Compensation on flow cytometer
In this protocol we used VersaComp Antibody Capture Bead Kit to correct spectral overlap by performing
fluorescence compensation following the manufacturer’s instruction.
Note: Compensation can be also done after sample acquisition with different flow cytometry data analysis
software (e.g., FlowLogic, FlowJo, etc.).
Quantification and Statistical Analysis
To assess the effector functions of human CD56dimCD16bright NK cells, we measured degranulation.
FlowJo software version 10.10.0 was used for the analysis of “fcs” files of flow cytometry data.
Expected Results
Identification of controls
In this protocol, we developed a control strategy for the correct identification of human CD56dimCD16bright NK cells in peripheral blood and the analysis of the degranulation
function of this subpopulation by flow cytometry.
Firstly, we started gating the flow data by using the Time gate based on the size (SSC: side scatter) and Time to identify acquisition issues (Figure 1A). Then we created lymphocytes gate based on their granularity (FSC: forward scatter) and size (SSC) (Figure 1B), and individual cells were selected based on FSC-A (area) and FSC-H (height) (Figure1C). Following these individual cells, the unstained control
(Figure 1D) and the FMO controls (Figure 1E-I) were determined, by selecting the fluorochrome of the marker to be measured according to lateral scattering (SSC). For a successful FMO control the percentage of cells counted should be < 1%.
Table1: Key materials and equipments
| REAGENT or RESOURCE | SOURCE |
| Antibodies | |
| Anti-human CD56-PE (clone MEM-188) | Biolegend, Koblenz, Germany |
| Anti-human CD16-FITC (clone 3G8) | Biolegend, Koblenz, Germany |
| Anti-human CD3-APC A-750 (clone UCHT1) | Biolegend, Koblenz, Germany |
| Anti-human CD107a APC (clone H4A3) | Biolegend, Koblenz, Germany |
| Annexin-V PB450A (lot: B380332) | Biolegend, Koblenz, Germany |
| Biological Samples | |
| Peripheral blood mononuclear cells (PBMCs) from healthy donors | Volunteers recruited at the National Blood Transfusion Centre (Centre National de transfusion sanguine,CNTS) |
| Reagents | |
| EDTA tubes of 10 mL | BD Vacutainer, K2E 18.0 mg, BD, Plymouth, UK |
| Ficoll-Paque Plus | Greiner Bio-One, GmbH, Germany |
| Fetal Calf Serum (FCS) | PAN Biotech, Aidenbach, Germany |
| Dimethylsulfoxide (DMSO) | Sigma Aldrich, St Louis, USA |
| Gentamycin | Life Technologies Corporation Grand Island, USA |
| RPMI 1640 medium | Gibco, California, USA |
| L-glutamine | Life Technologies Limited, Paisley, UK |
| Penicillin-Streptomycin | Life Technologies Corporation Grand Island, USA |
| Phosphate Buffered Saline (PBS) | Gibco, Thermo Fisher Scientific |
| Trypan Blue Solution | Life Technologies Corporation, Grand Island, USA |
| VersaComp Antibody Capture Bead Kit | Beckman Coulter, Brea, California, USA |
| Software and Algorithms | |
| FlowJo™ Version 10.10.0 | FlowJo LLC |
| GraphPad Prism v9.0 | Prism-GraphPad |
| Other (Equipement, consummables) | |
| Analyzer flow cytometer (Cytoflex) | Beckman Coulter Technology, Suzhou, China |
| Falcon® conical 50 mL centrifuge tubes | Falcon Greneir Bio one, Frickenhausen, Germany |
| Falcon conical 15 mL Centrifuge Tubes | Falcon Greneir Bio one, Frickenhausen, Germany |
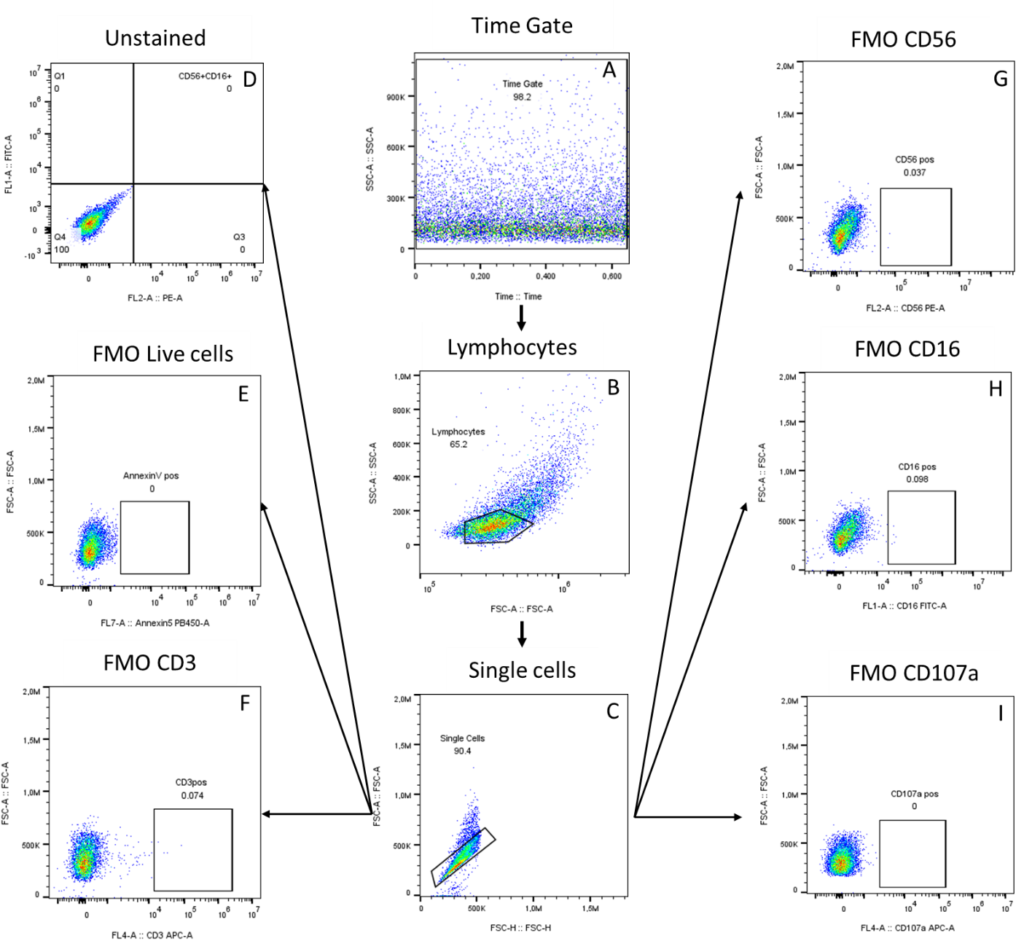
Figure 1: FMO Controls gating strategy. The arrows indicate the gating of controls A) Identification of acquisition issues in the Time gate based on the SSC-A surface area and Time B) Selection of lymphocytes cells based on FSC (granularity) and SSC (size); C) Single cells based on FSC-A (area) and FSC-H (height); D) Unstained; E) FMO Annexin V; F) FMO CD3; G) FMO CD56; H) FMO CD16; I) FMO CD107a
Note 1: The time gate may have debris in the data due to air or obstructions during acquisition. In this case it is recommended to create the time gate by eliminating these problematic areas.
Note 2: A second purification of individual cells can also be performed by selecting individual cells according to SSC-A surface area and SSC-H height by side-scattering.
Functional analysis of NK cells: degranulation assay
Once the controls were determined, CD56dimCD16bright NK cells were correctly identified, and the effector functions of this subset were assessed. We started gating the flow data by using the Time gate based on the size (SSC: side scatter) and Time to identify acquisition issues (Figure 2A). We created lymphocyte gates based on their forward scatter (FSC) and side scatter (SSC) parameters (Figure 2B). Then, individual cells were selected based on FSC-A (area) and FSC-H (height) (Figure 2C). Then an exclusion channel (Annexin-V viability dye) was included in our control strategy to specifically study viable cells (Figure 2D). From the viable Annexin-V- (negative) cells anti-CD3 monoclonal antibody was included to exclude T cells (Figure 2E). Thus, the NK cells were identified for the non-T cells population, based on the expression of the surface markers CD56 and CD16 (Figure 2F) and different subsets were selected from CD56pos and CD16pos (Figure 2G). To measure the degranulation of the CD56dimCD16pos NK cells subset, we included anti-CD107a monoclonal antibodies, and degranulated NK cells expressing CD107a marker, were identified from CD56dimCD16pos (Figure 2H).
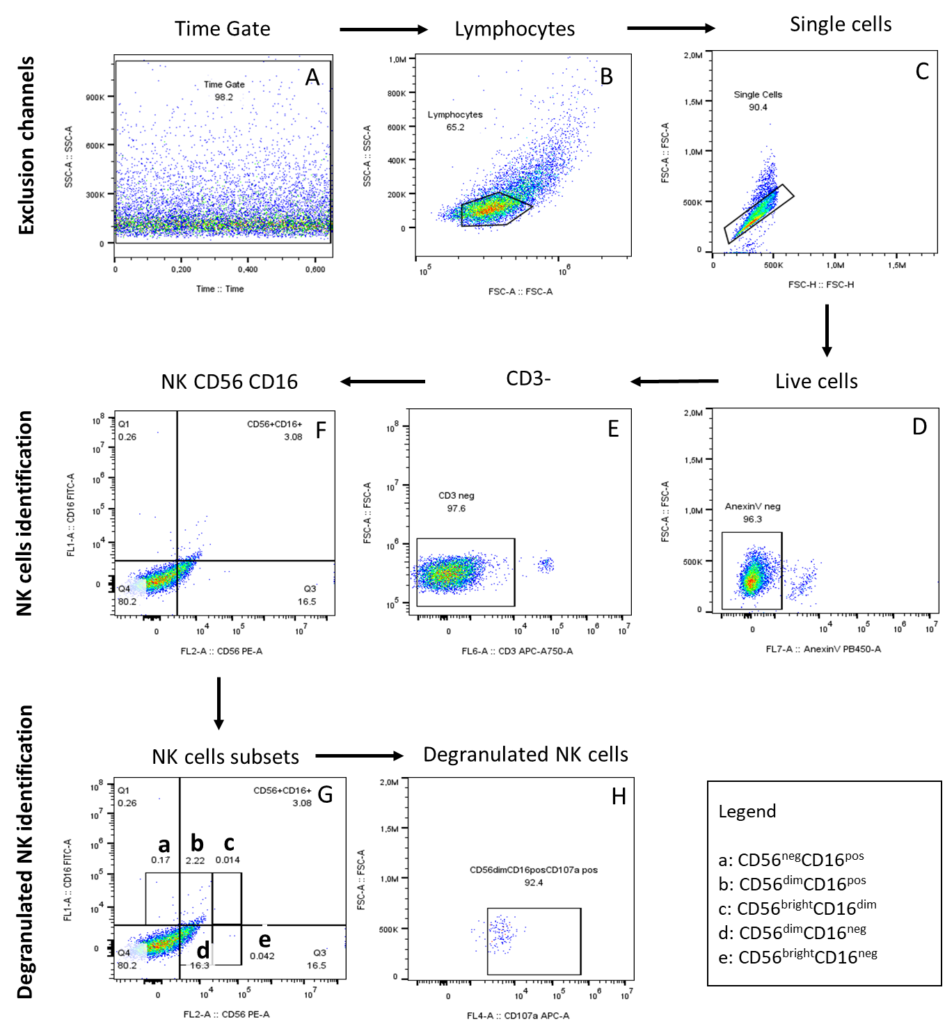
Figure 2: Gating strategy of degranulated NK cells identification. The arrows show the gating steps for the expression of CD107a on the NK cells. A) Identification of acquisition issues in the Time gate B) Selection of lymphocytes based on FSCA and SSC-A; C) Single cells based on FSC-A and FSC-H forward scatter; D) Live cells E); CD3neg cells F); NK cells based on the CD56 and CD16 markers G); NK subsets cells a) CD56negCD16pos b) CD56dimCD16pos c) CD56brightCD16dim d)CD56dimCD16neg e) CD56brightCD16neg H); Degranulated NK cells expressing CD107a identified from CD56dimCD16possubset.
Limitations
NK cells play an important role in innate immune responses. In this protocol we describe a method to identify different NK cells subsets and their degranulation function. This protocol uses PBMCs for the identification of NK cells, and this identification included only anti- Annexin-V and anti-CD3 exclusion channel for the non-viable cells and T cells respectively, since NK cells are CD3- (negative) (10). Although T cells are excluded, some B cells may interfere with the identification of NK cells. However, to ensure the
purity of NK cells we propose to include a B cell exclusion channel with anti-CD19 and anti-CD14 monoclonal antibodies. Furthermore, annexin-V is an apoptotic cells marker and cannot be used to identify other type of dead cells. We propose then to use a complete viability marker for the analysis of cell viability such as LIVE/DEAD™ Fixable Blue Dead Cell Stain Kit.
The regulated expression of degranulated NK cells may be due to direct staining of NK cells after isolation. However, the effector function of NK cells underlies their activation through IL-2 or other stimuli (11). Thus, cells stimulation with Phorbol 12-myristate 13-acetate (PMA), IL-2+IL-2+IL-18 or antibodies for 24 or 48 hours, depending on the type of infection studied, would increase the percentage of degranulated cells. Nevertheless, the gating strategy proposed in this protocol enables NK cells and their
degranulation functions to be properly identified.
Conflict of interest
The authors declared no conflict of interest.
Author’s contribution
Simfele Hombamane Christelle: Conceptualization, Formal Analysis, Writing Original Draft, Methodology;
Nadjir Koboyo Liza: Enrolment; Katawa Gnatoulma: Conceptualization, Investigation, Data curation, Project Administration, Funding Acquisition, Methodology, Resources, Supervision; Kolou Malewe: Validation.
References
- Caligiuri MAJB, The Journal of the American Society of Hematology. Human natural killer cells. 2008;112(3):461-9.
- Thiruchelvam U, Wingfield M, O’Farrelly CJAJoRI. Natural killer cells: key players in endometriosis.
2015;74(4):291-301. - Abel AM, Yang C, Thakar MS, Malarkannan SJFii. Natural killer cells: development, maturation, and clinical utilization. 2018;9:1869.
- Yu J, Freud AG, Caligiuri MAJTii. Location and cellular stages of natural killer cell development. 2013;34(12):573
- Flórez-Álvarez L, Hernandez JC, Zapata WJFii. NK cells in HIV-1 infection: from basic science to vaccine strategies. 2018;9:2290.
- Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends in immunology. 2001;22(11):633-40.
- Mavilio D, Lombardo G, Benjamin J, Kim D, Follman D, Marcenaro E, et al. Characterization of CD56–/CD16+natural killer (NK) cells: A highly dysfunctional NK subset expanded in HIV-infected
2005;102(8):2886-91. viremic individuals. - Prada N, Antoni G, Commo F, Rusakiewicz S, Semeraro M, Boufassa F, et al. Analysis of NKp30/NCR3 isoforms in untreated HIV-1-infected patients from the ANRS SEROCO cohort. 2013;2(3):e23472.
- Della Chiesa M, Marcenaro E, Sivori S, Carlomagno S, Pesce S, Moretta A, editors. Human NK cell response to pathogens. Seminars in immunology; 2014: Elsevier.
- Moretta A, Bottino C, Pende D, Tripodi G, Tambussi G, Viale O, et al. Identification of four subsets of human CD3 CD16+ natural killer (NK) cells by the expression of clonally distributed functional surface molecules: correlation between subset assignment of NK clones and ability to mediate
specific alloantigen recognition. The journal of experimental medicine. 1990;172(6):1589-98. - Pahl JH, Koch J, Götz J-J, Arnold A, Reusch U, Gantke T, et al. CD16A activation of NK cells promotes NK cell proliferation and memory-like cytotoxicity against cancer cells. Cancer immunology research. 2018;6(5):517-27.
